Introduction
The achievement of personalized and precision medicine demands not only massive multi-modal analysis of medical records, but also detailed international healthcare data to build accurate predictive models properly stratified to different sub-populations. Data integration is particularly crucial for rare diseases, where data are scarce, extremely heterogeneous, and disseminated in many repositories around the globe. In this study, we tested how well the use of a high-level semantic model, Biolink,1 was able to support integration of data from two independent sources: Critical Path Institute’s (C-Path) Rare Disease Cures Accelerator Data and Analytics Platform (RDCA-DAP) and European Joint Programme on Rare Diseases (EJP-RD)—when both sources used Findable, Accessible, Interoperable, and Reusable (FAIR) Data Principles to represent their data, but each used an independently developed data model.2
Background
Healthcare data, like most scientific data, is spread over many formats and repositories. This alone makes them challenging to integrate, but the data also have features, such as being highly privacy-sensitive, that dramatically inhibit integration. Within life and health sciences, interoperability projects that focused on machine-actionability began in the late 1990s, and some came to fruition in the early 2000s. Some approaches pushed the interoperability problem on to the data owner, trying to enforce harmonization at-source, such as caBIO and TAPIR.345 caBIO created an interface standard for the cancer genetics/genomics community that all participating organizations should implement; whereas TAPIR, for the biodiversity community, pursued a query language that all sites should respond to. With respect to more general-purpose approaches, myGrid and BioMoby were both Web-services-based interoperability projects that used semantic annotations of Web service interfaces, and in the case of BioMoby, an ontology-driven eXtensible Markup Language (XML) Schema to harmonize data structures.678 SADI and SSWAP used the Semantic Web technologies of Resource Description Framework (RDF) and Web Ontology Language (OWL) to annotate Web Service interface definitions and required that all data passed between their participating services be represented as ontologically grounded RDF.910 Notably, none of the approaches mentioned thus far have attempted to achieve integration purely at the level of the data itself. This is at least in part due to legacy data formats being either unstructured or structured (for the purpose of sharing) in the form of XML documents, XML being described as “far and away the most complex data model ever proposed” and “seriously flawed”.11
The now more widespread use of Semantic Web technologies should, in principle, make it easier to attempt integration at the level of the data itself, rather than via Web services, tightly defined query interfaces, or explicit sharing of schemas or models. Nevertheless, successful examples are still lacking. This manuscript describes our attempt to achieve interoperability between two non-coordinating data sources purely through the introduction of shared semantic mapping.
The FAIR principles are a set of guidelines for publication of data and metadata that enhance the ability of datasets to be discovered and processed by machines. The principles focus largely on data reuse, which implicitly requires interoperability.2 While the FAIR Principles focus largely on metadata, the information that describes the content of data, they also encourage the reformatting of the data into machine-readable syntaxes, with machine-readable semantics (most often implemented using RDF/OWL).
FAIR forms the basis of the EJP-RD Virtual Platform, where several dozen participating rare disease registries and biobanks are being prepared for integrative queries via transformation of their contents into FAIR data formats.12 As an initial target for interoperability between EJP-RD datasets, 16 Common Data Elements (CDEs) were selected, as defined by the European Platform on Rare Disease Registration.13 A core FAIR semantic model based on Semantic Science Integrated Ontology (SIO) was designed to act as a scaffold for these Common Data Elements, enabling model reusability and thus simplifying query.14 All CDE data within the EJP-RD are transformed into RDF that adheres to this core model.
FAIR principles also guide the development of the RDCA-DAP.151617 RDCA-DAP provides a neutral environment for industry, academia, regulators, and other government agencies to work together to accelerate and de-risk the medical product development process in rare diseases. RDCA-DAP integrates existing datasets from various sources, including clinical trials, patient registries, preclinical data, natural history studies, and electronic health records. Much of the data in RDCA-DAP originates from clinical trials and is therefore already standardized, using standards from the Clinical Data Interchange Standards Consortium (CDISC). CDISC is a standards development organization that maintains and develops a suite of data standards that encompass data acquisition through analysis.18 Collected data are represented by the CDISC Study Data Tabulation Model (SDTM), a common data model required for clinical trial data submission to several regulatory agencies.19 The model provides a standard for the representation of collected data into domains of similar biomedical concepts, such as demographics, laboratory tests, or concomitant medications. Because of its use in regulatory activities, SDTM is strongly aligned with clinical trial (CT) data. The growing importance of real-world data has raised interest in integrating SDTM-formatted CT data with other data types, such as patient registries and electronic health records.20
To integrate trial data with other data types and support cross-disease data exploration, an initiative is underway to build semantically grounded models that represent RDCA-DAP data in OWL.
Biolink is a high-level biological domain data model used to represent biomedical entities and the relationship between them. Entities are annotated with ontological terms to semantically ground their meaning.1 Although Biolink was developed using LinkML (a YAML-based schema modelling language),21 it has been translated into multiple formats, including an OWL version available through BioPortal.22 Biolink has links to external ontologies, and it has been used in a variety of health registries and projects, such as the KG-COVID-19 project.23
Methods
Coordination was undertaken through weekly meetings by a group with a range of differing expertise, including Semantic Web, Linked Data, and HealthCare data management. The group was formed from representatives of C-Path and EJP-RD organizations, with the larger goal of achieving data interoperability via federation between both organizations’ datasets.
For initial analysis, each organization provided an existing dataset about patients with polycystic kidney disease (PKD). The C-Path dataset consists of aggregated data from multiple studies that was gathered by the PKD Outcomes Consortium.24 These data have been used to develop CDISC data standards for PKD and to support the Food and Drug Administration (FDA) and European Medicines Agency (EMA) qualifications of Total Kidney Volume as an imaging biomarker for drug development tools. The already anonymized dataset was further protected for this study by synthesizing values for laboratory test results using the synthpop R package.25 The EJP-RD dataset consists of mock data spanning three clinical information domains, described in Table 1. These domains were selected from both datasets as the initial targets for federation because they contain semantically similar entities that appear in both the C-Path and EJP-RD semantic data models and datasets. These entities were then mapped to the Biolink model (Table 2). National Cancer Institute Thesaurus (NCIT) terms were used for specific fields (e.g., sex, laboratory test names), because study data tabulation model (SDTM) controlled terminologies are already mapped to NCIT, and EJP-RD had already chosen the NCIT OBO version as the reference ontology for several domains.26
Selected clinical information types present in both datasets.
| Domain | C-Path | EJP-RD |
|---|---|---|
| Birthdate/Age patient information | Age as an integer | ISO 8601 compliant date string |
| Sex patient information | Sex label as a string (F, M) mapped to NCIT terms for Female and Male | NCIT term for Sex (Female, Male, Undetermined, or Unknown) and Sex label as a string |
| Laboratory data measurements | • Laboratory test name (e.g., Leukocytes), category (e.g., hematology), and specimen type (e.g., blood) • Numerical result and standard ranges in both original and standard units • Study day of lab test • Associated subject visit • Associated specimen collection procedure |
• Procedure defined as Quantitation or Estimation • Materials tested input • Target molecular or compound measured • Output measurement value and its unit • ISO 8601 compliant date of measurement procedure |
Mapping of similar conceptual entities between Biolink, C-Path, and EJP-RD.
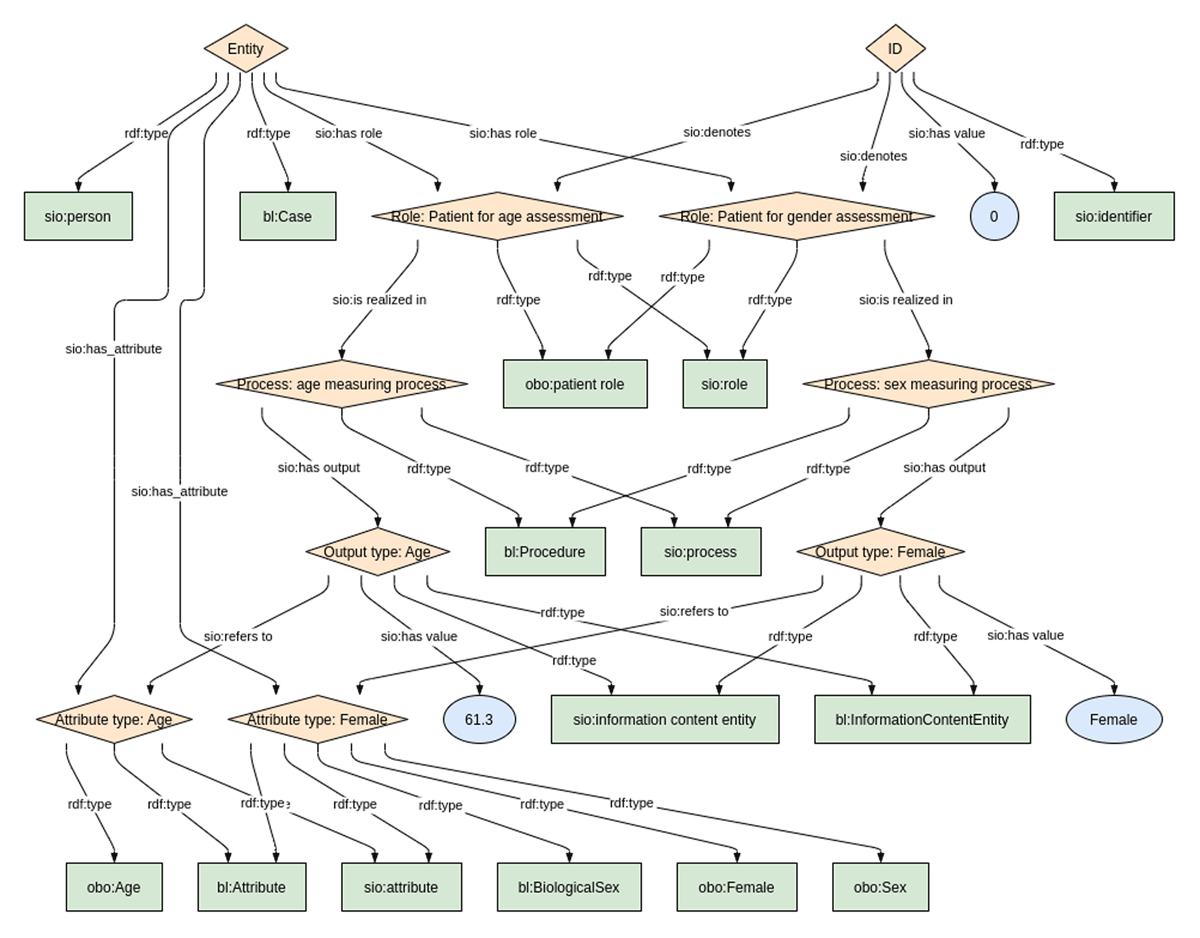
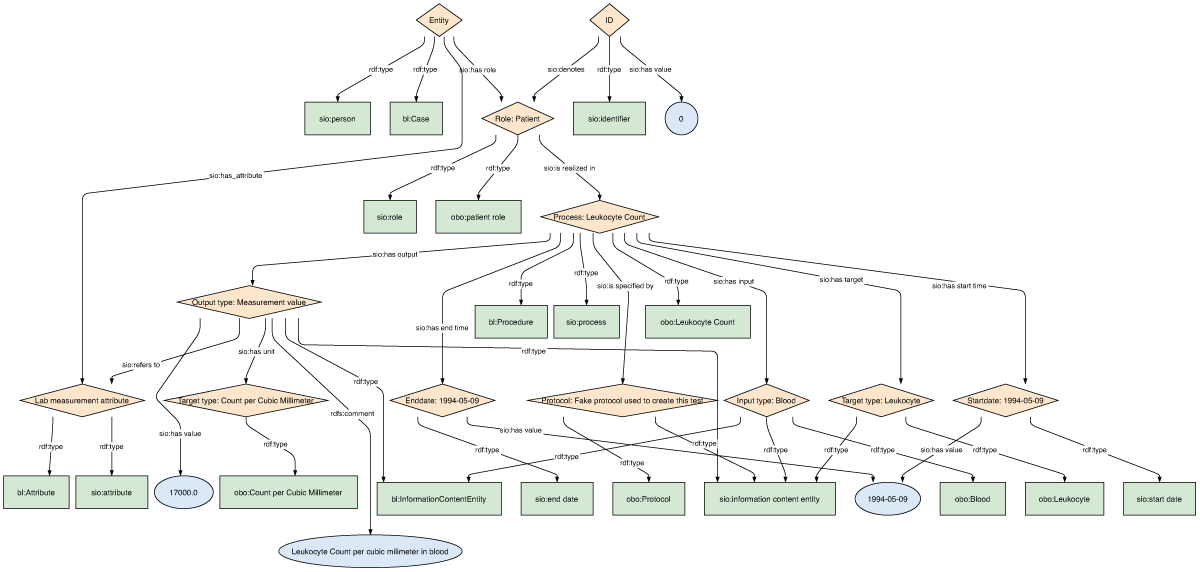
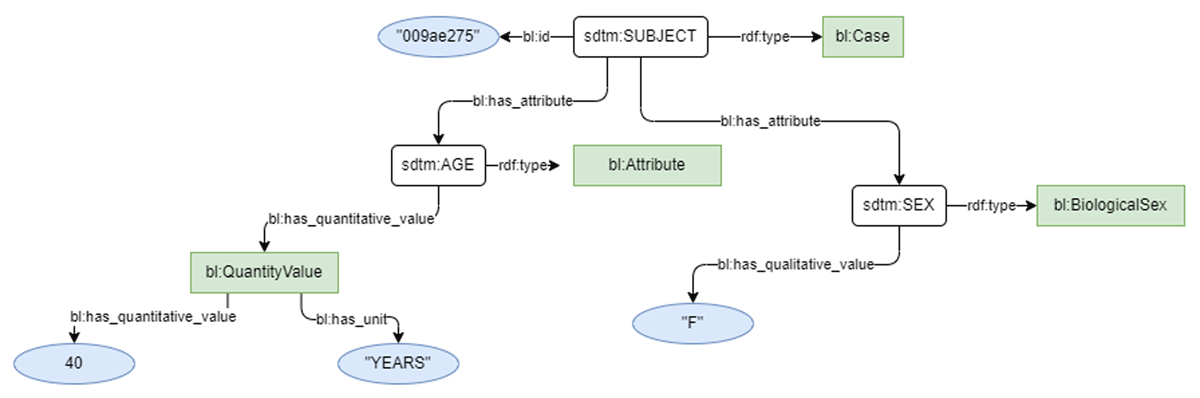
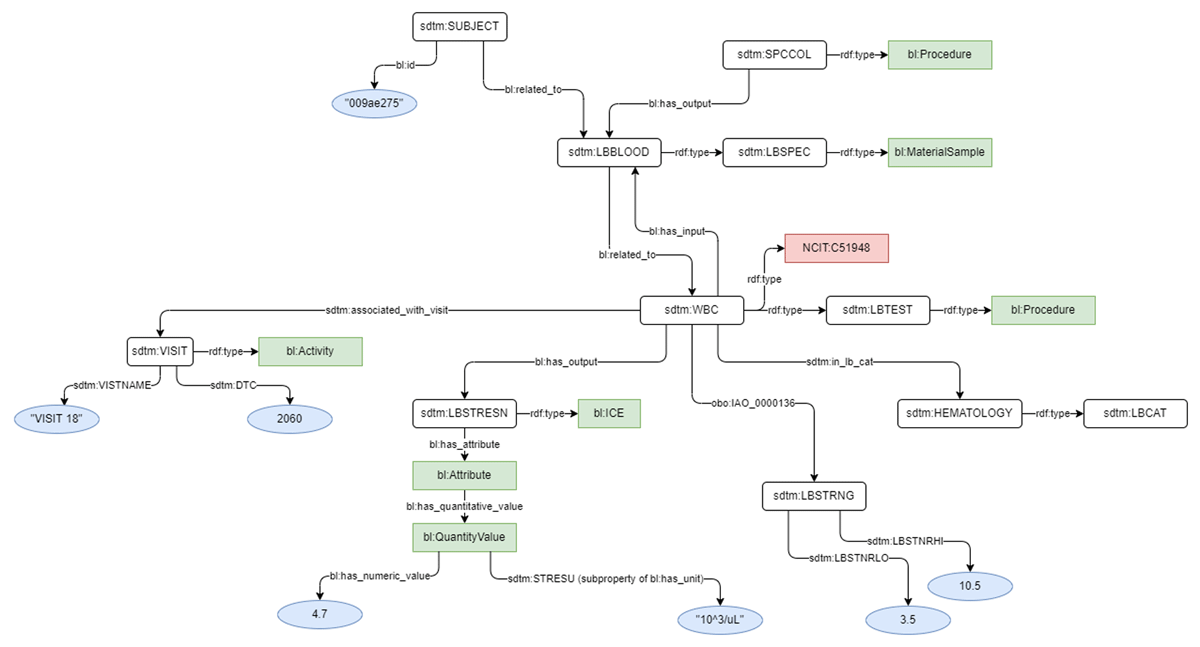
To execute the study, the shared Biolink concept URLs were added into both C-Path and EJP-RD data models to harmonize the type of each shared concept, but otherwise the source models were left unchanged. Models for Personal Information and Leukocyte Count are shown for both the EJP-RD (Figures 1 and 2) and C-Path (Figures 3 and 4). Each project generated RDF data using YARRRML.27 C-Path ontology, source data, and RDF conversion code are available on GitHub.28 Both C-Path and EJP-RD data are available on a server.29
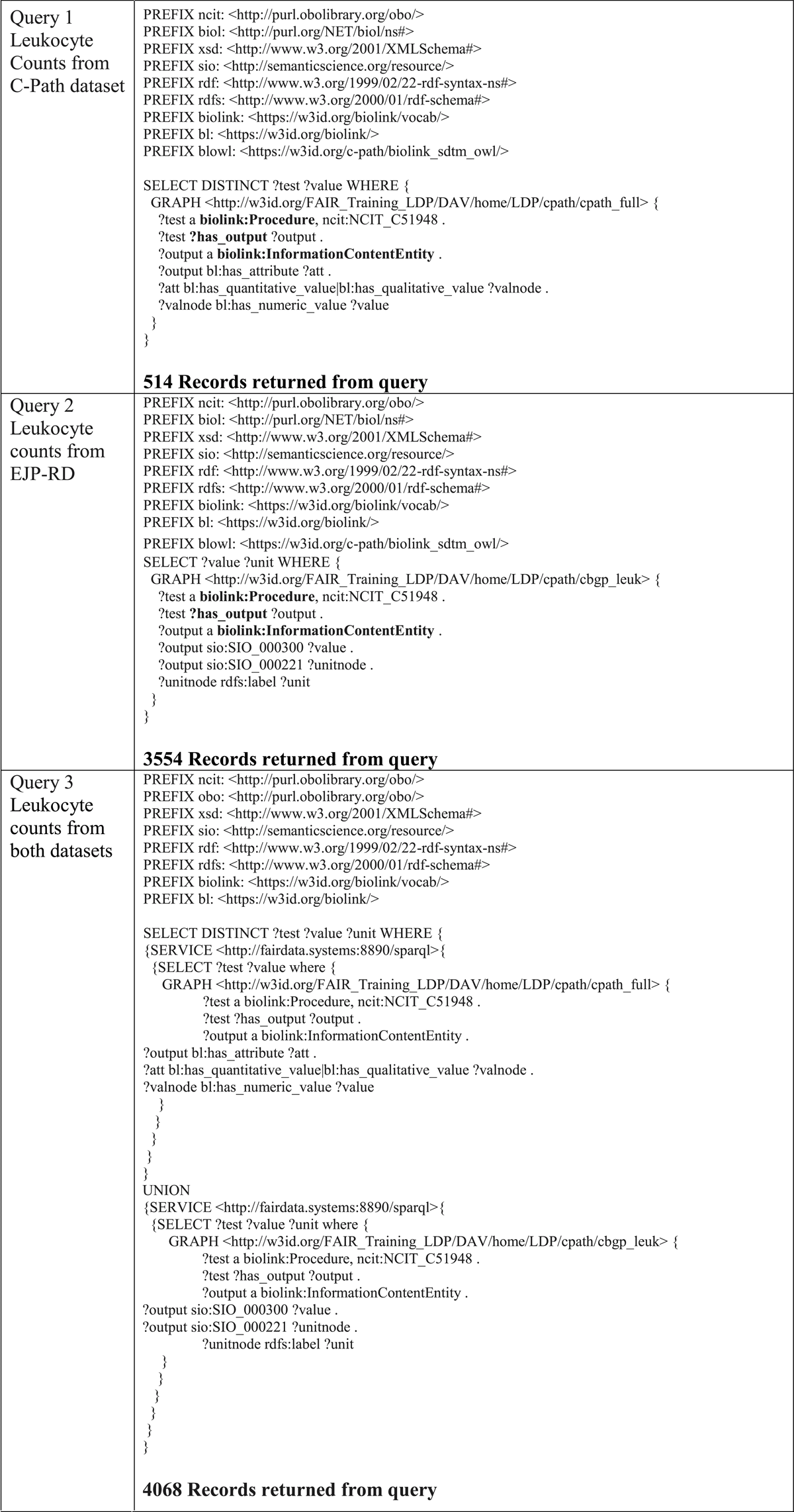
RDF data from each project were loaded into separate named graphs, such that they could be queried independently. The SPARQL query language was selected as the tool used to explore interoperability between the two datasets. Data can be queried at our SPARQL endpoint by copying or modifying the queries in Table 3.30 A critical feature of SPARQL is that all aspects of a data schema, both entities and relationships, can be represented as variables. This is relevant because the EJP-RD models utilize SIO to describe concept-to-concept relationships, while C-Path uses a customized extension of the Biolink model. Therefore, by leveraging this feature of SPARQL, we attempted to construct near-identical queries over both data models by leaving, as variables, all aspects of the model that are not shared—that is, the queries use only the shared Biolink-typed nodes, leaving all disparate aspects of the underlying models as query variables.
Results
To demonstrate that we have achieved interoperability, SPARQL queries were constructed, with exemplar queries shown in Table 3. Query 1 extracts leukocyte count from the C-Path dataset. Query 2 extracts leukocyte count from the EJP-RD dataset. Query 3 uses the SPARQL SERVICE clause to show how a federated query would be constructed over multiple registries; however, in this case, the data is hosted in two separate graphs on the same server.
Discussion
Our results demonstrate that independent mapping of datasets from two distinct data models to an upper-level schema, such as Biolink, offers a starting point of interoperability. In principle, the components of the shared model, such as subjects or patients (e.g., the Case class in Biolink) and their attributes (e.g., the Attribute class in Biolink), provided a detailed-enough structure that independent mapping to these elements provided the means of constructing queries that retrieved records from both datasets using only the shared set of Uniform Resource Identifiers (URI). This is exemplified by the bolded features in Queries 1 and 2 in Table 3, showing that we can at least partially anchor the query around the shared typed components while leaving the sub-structure of the disparate data models as a query variable. The further extension of this interoperability will require additional collaboration to harmonize how more general shared concepts, such as procedures and activities, relate to the underlying literal values in each dataset.
This early-stage investigation had some limitations. Several actions were taken that, in a realistic scenario, would not be possible. For example, adding an additional type node into the EJP-RD data model was only possible because we are the custodians of that data and, thus, were able to create a separate dataset that carried this additional information. This was done to set a baseline for how much interoperability we might expect from this mapping effort. Nevertheless, this direct manipulation of the data is not strictly necessary to achieve the goals pursued here. For example, the mapping in Table 2 could be captured in RDF and published independently. This would enable a query to be constructed that utilized the mapping data as a third SPARQL SERVICE (i.e., extending Query 3 to make a third SERVICE call to the mapping dataset). This alternative approach would also solve another, more subtle, problem that arises by adding Biolink types directly into the data. That is, Biolink is a schema, not an ontology. In RDF, the type property has a strict semantic meaning and should only be used to categorize a concept based on an ontology term. Thus, by adding Biolink types into the EJP-RD dataset, we render it unusable for the purpose of logical reasoning. Because the C-Path model was built using the OWL version of Biolink, this exception does not apply to the C-Path data, but the limited semantics of Biolink do limit the reasoning that is possible.
Other identified limitations, however, cannot be so easily resolved. One example is the use of age in the personal information from one dataset versus date of birth in the personal information of the other (not shown). While it would be possible to extract both data types using SPARQL (e.g., by simply creating a query that gathers all personal information), it is not possible to harmonize the data within the query itself, thus requiring some degree of post-processing of the results. This, however, is not atypical in clinical research or meta-analyses, and thus we still consider the gain in query power of FAIR data to be useful. Other limitations were observed that we will not detail here, such as inconsistent use of units across datasets, data cleaning and reformatting that must be done before conversion to RDF, and the lack of standardized vocabularies for many domains of clinical data. Some can be overcome by enhancing the semantic content within both datasets.
FAIR representations of legacy data require additional curation work, but in our experience, they are generally not more time-consuming than mapping data to less FAIR standards (e.g., that do not use standardized vocabularies or include URLs for their terminology). It is difficult to estimate the total person hours required for us to carry out this project, which took place over about three months, because the most time-consuming parts—building the semantic models and (for C-Path) mapping data to SDTM—were already done. During the three-month project period, our teams met online weekly to make sure we understood each other’s models and our mappings to Biolink, because all the models are new and not yet well-known. However, an end goal is to allow independent mappings for improved interoperability.
Conclusions and Future Directions
This is an early report of an initiative to enable interoperability between two large international clinical data sharing initiatives: C-Path and EJP-RD. Biolink was chosen as the enabling technology to establish FAIR Data, and in the limited exploratory datasets created for this study, it was found to enable some degree of cross-compatible federated query over the two datasets. A CDISC contribution that would speed up this and similar work would be the creation of globally unique, permanent, resolvable identifiers for SDTM terms (both standard variables and value terminologies). Such IDs would allow direct reuse of SDTM in knowledge graphs, without having to create duplicate terms in an ontology. The mapping of NCIT terms to CDISC terminology is a step in this direction that allowed us to reuse NCIT for some, and work undertaken in the CDISC 360 initiative is highly relevant in this context.31 We are examining alternative mechanisms to enhance interoperability while reducing the degree to which any participating dataset must modify its contents, such as publishing an external mapping between the two models (mentioned in the discussion) and better semantic encoding within each dataset (e.g., replacing the sex values in C-Path data with NCIT concepts). We will continue to provide access to semantic models and analysis and data transformation code via public repositories. Data will continue to be available through C-Path’s RDCA-DAP and will be delivered via the EJP-RD Virtual Platform when it goes live.
Acknowledgements
PAM, NB, RC, and MDW are supported by funding from the European Union’s Horizon 2020 research and innovation programme under the EJP RD COFUND-EJP N° 825575. IB, EH, DO, and RLW are supported by the Critical Path Institute, which is supported by the FDA of the U.S. Department of Health and Human Services (HHS), and is 54.2% funded by the FDA/HHS, totaling $13,239,950, and 45.8% funded by non-government source(s), totaling $11,196,634. The contents are those of the author(s) and do not necessarily represent the official views of, nor are an endorsement by, FDA/HHS or the U.S. Government.
Competing Interests
The authors have no competing interests to declare.
Author Contributions
Pablo Alarcón-Moreno and Ian Braun contributed equally.
References
1. Biolink Model. https://biolink.github.io/biolink-model/. Accessed February 2, 2022.
2. Wilkinson MD, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016 Mar 15; 3: 160018. Erratum in: Sci Data. 2019 Mar 19; 6(1): 6. PMID: 26978244; PMCID: PMC4792175. DOI: http://doi.org/10.1038/sdata.2016.18
3. Covitz PA, Hartel F, Schaefer C, et al. caCORE: A common infrastructure for cancer informatics. Bioinformatics. 2003; 19(18): 2404–2412. DOI: http://doi.org/10.1093/bioinformatics/btg335
4. Phillips J, Chilukuri R, Fragoso G, Warzel D, Covitz PA. The caCORE Software Development Kit: Streamlining construction of interoperable biomedical information services. BMC Med Inform Decis Mak. 2006; 6(1): 1–16. DOI: http://doi.org/10.1186/1472-6947-6-2
5. TAPIR–TDWG Access Protocol for Information Retrieval. http://tdwg.github.io/tapir/docs/tdwg_tapir_specification_2010-05-05.html. Accessed January 28, 2022.
6. Stevens RD, Robinson AJ, Goble CA. myGrid: Personalised bioinformatics on the information grid. Bioinformatics. 2003; 19(suppl_1): i302–i304. DOI: http://doi.org/10.1093/bioinformatics/btg1041
7. Wilkinson MD, Links M. BioMOBY: An open source biological web services proposal. Brief Bioinform. 2002; 3(4): 331–341. DOI: http://doi.org/10.1093/bib/3.4.331
8. Wilkinson MD, Senger M, Kawas E, et al. Interoperability with Moby 1.0—It’s better than sharing your toothbrush! Brief Bioinform. 2008; 9(3): 220–231. DOI: http://doi.org/10.1093/bib/bbn003
9. Wilkinson MD, Vandervalk B, Mccarthy L, Wilkinson M. The Semantic Automated Discovery and Integration (SADI) Web service Design-Pattern, API and Reference Implementation. Nat Preced 2011. October 2011; 1–1. DOI: http://doi.org/10.1038/npre.2011.6550.1
10. Gessler DDG, Schiltz GS, May GD, et al. SSWAP: A simple semantic web architecture and protocol for semantic web services. BMC Bioinformatics. 2009; 10(1): 309. DOI: http://doi.org/10.1186/1471-2105-10-309
11. Hellerstein JM, Stonebraker M. Readings in database systems. 2005:865. https://mitpress.mit.edu/books/readings-database-systems-fourth-edition. Accessed February 2, 2022.
12. European Joint Programme on Rare Diseases. 2022. What is the Virtual Platform. https://www.ejprarediseases.org/what-is-it/. Accessed April 20, 2022.
13. Set of common data elements for rare diseases registration. https://eu-rd-platform.jrc.ec.europa.eu/sites/default/files/CDS/EU_RD_Platform_CDS_Final.pdf. Accessed June 30, 2021.
14. Dumontier M, Baker CJO, Baran J, et al. The semanticscience integrated ontology (SIO) for biomedical research and knowledge discovery. J Biomed Semantics. 2014; 5(1): 1–11. DOI: http://doi.org/10.1186/2041-1480-5-14
15. C-Path RDCA-DAP Portal. https://portal.rdca.c-path.org/. Accessed February 16, 2022.
16. Critical Path Institute. https://c-path.org/. Accessed December 15, 2021.
17. Woosley RL, Myers RT, Goodsaid F. The Critical Path Institute’s Approach to Precompetitive Sharing and Advancing Regulatory Science. Clin Pharmacol Ther. 2010; 87(5): 530–533. DOI: http://doi.org/10.1038/clpt.2010.27
18. Cdisc.org. 2022. Global Regulatory Requirements | CDISC. https://www.cdisc.org/resources/global-regulatory-requirements. Accessed April 18, 2022.
19. Cdisc.org. 2022. SDTM | CDISC. https://www.cdisc.org/standards/foundational/sdtm. Accessed April 18, 2022.
20. Arlett P, Kjær J, Broich K, Cooke E. Real-World Evidence in EU Medicines Regulation: Enabling Use and Establishing Value. Clin Pharmacol Ther. 2022; 111(1): 21–23. DOI: http://doi.org/10.1002/cpt.2479
21. YAML Ain’t Markup Language (YAMLTM) revision 1.2.2. https://yaml.org/spec/1.2.2/. Accessed February 2, 2022.
22. Noy NF, Shah NH, Whetzel PL, et al. BioPortal: Ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009; 37(suppl_2): W170–W173. DOI: http://doi.org/10.1093/nar/gkp440
23. Reese JT, Unni D, Callahan TJ, et al. KG-COVID-19: A Framework to Produce Customized Knowledge Graphs for COVID-19 Response. Patterns. 2021; 2(1). DOI: http://doi.org/10.1016/j.patter.2020.100155
24. PKD | Critical Path Institute. https://c-path.org/programs/pkd/. Accessed February 17, 2022.
25. synthpop: Generating Synthetic Versions of Sensitive Microdata for Statistical Disclosure Control. https://CRAN.R-project.org/package=synthpop
26. NCI Thesaurus OBO Edition. https://obofoundry.org/ontology/ncit.html. Accessed February 17, 2022.
27. YARRRML. https://rml.io/yarrrml/spec/. Accessed February 2, 2022.
28. GitHub – criticalpathinstitute/biolink_sdtm_owl: An ontology for a proof of concept mapping of PKD data in the SDTM format to the Biolink Model. https://github.com/criticalpathinstitute/biolink_sdtm_owl. Accessed February 17, 2022.
29. WebDAV Repository. http://fairdata.systems:8890/DAV/home/LDP/cpath/. Accessed February 17, 2022.
30. Virtuoso SPARQL Query Editor. http://fairdata.systems:8890/sparql. Accessed February 17, 2022.
31. CDISC 360. https://www.cdisc.org/cdisc-360