Introduction
The US National Cancer Institute’s Enterprise Vocabulary Services (NCI-EVS) provides terminologies, technological solutions, and analysis services to accurately code and share biomedical research data, clinical information, and public health information.1 NCI-EVS provides the foundational layer for the National Cancer Institute’s (NCI’s) semantic infrastructure through the NCI Thesaurus (NCIt) and NCI Metathesaurus (NCIm) biomedical coding terminologies.2,3,4,5 NCI-EVS engages in many national and international partnerships to develop, license, publish, and distribute terminology, and develops software tools that support data sharing and semantic interoperability. Prominent stakeholders include the US Food and Drug Administration (US FDA) and the Clinical Data Interchange Standards Consortium (CDISC). NCI-EVS directly partners with both organizations to develop and maintain their controlled terminologies in the freely available, non-license restricted terminology environment of the NCIt.
Since 2017, CDISC data standards, including CDISC Controlled Terminologies (CT), have been required for use in electronic data submissions to the US FDA and the Japanese Pharmaceuticals and Medical Devices Agency (PMDA).6,7 Additionally, the CDISC data standards are currently preferred by the Chinese National Medical Products Administration (NMPA, formerly the CFDA) and recommended by the European Medicines Agency (EMA).8,9 CDISC CT, along with other prominent biomedical coding terminologies are integrated into regulatory Data Standards Catalogs and Study Data Technical Conformance Guides.10,11
A recently published framework document from the US FDA indicates that Real-World Evidence (RWE) generated from Real-World Data (RWD) will support future regulatory decisions regarding the safety and effectiveness of drugs and biological products.12 Additionally, the EMA is exploring ways in which data derived from randomized controlled trials (RCT) and RWD can be exploited for safety and efficacy decisions.13 However, RWD is generated from multiple systems and sources that use disparate data standards (if any), vocabularies, and coding systems. RWD is therefore generally considered to be less structured than regulated clinical research data.
NCI-EVS Services and Products
NCI-EVS provides an array of technology products and services to support the semantic infrastructure of NCI and other collaborating organizations. NCI-EVS services include the following: subject matter expertise; definition writing and analysis; terminology coding, tagging, and subset bundling; terminology publication, maintenance, and versioning; mapping of NCIt content to other controlled vocabularies and data standards; and content addition and enrichment based on evolving scientific discoveries and terminology requests from users. NCI-EVS domain experts follow good terminology practice principles in the development of terminology and ontology content.5,14,15 NCI-EVS maintains a Term Suggestion form maintains a Term Suggestion form that enables direct interaction of the user community with NCI-EVS domain experts.16 This web form facilitates requests for application facilitates requests for new terms or changes to existing terms and provides a mechanism for expert consulting services and knowledge transfer.
NCI Thesaurus (NCIt)
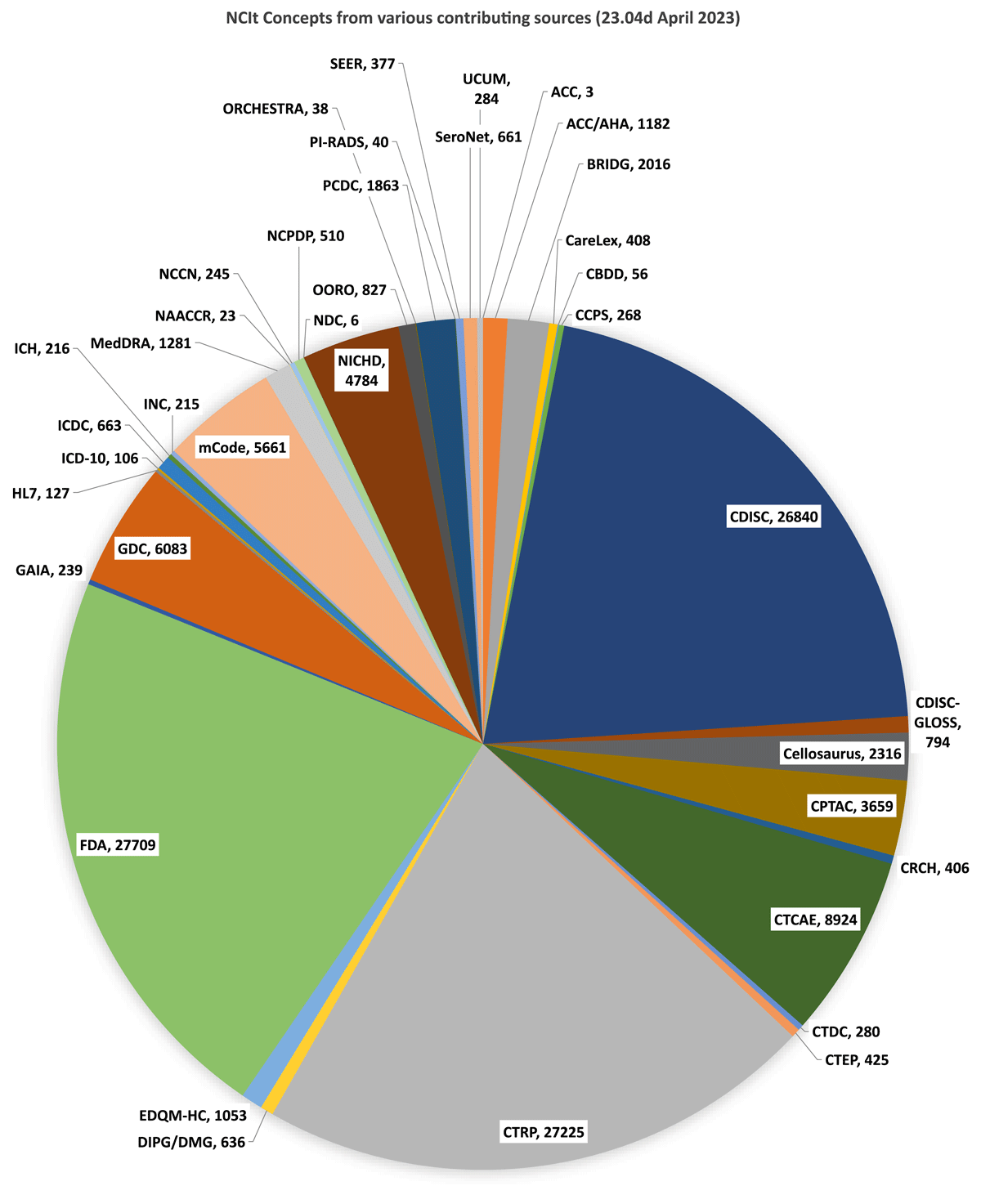
The NCI Thesaurus (NCIt) is a free, non-license restricted biomedical coding terminology that contains over 176 000 concepts that span basic research, clinical care, translational research, healthcare activities, public information, and administrative endeavors. It is an internationally recognized biomedical coding standard, which is organized into a richly structured, description logic-based hierarchy that has more than 400 000 modeled relationships between concepts. NCIt expansion is generally driven by the needs of the NCI, the US National Institutes of Health (NIH), and term source partners. However, anyone can suggest an addition or a change to existing concepts in NCIt through the Term Suggestion form.16 In all, there are 50 source contributors tagged in NCIt concepts (Figure 1). A public browser, Application Programming Interface (API), and monthly subset report generation in multiple file formats ensures a high level of accessibility to the content. NCIt content is managed through an NCI-EVS-specific instance of Protégé, an ontology editor developed by Stanford University.17
Concept numbers tagged in NCIt per Contributing Source organization. [ACC (American College of Cardiology); ACC/AHA (American College of Cardiology/American Heart Association); BRIDG (Biomedical Research Integrated Domain Model Group); CareLex (CareLex electronic Trial Master File Terminology); CBDD (NCI’s Chemical Biology and Drug Development); CCPS (Childhood Cancer Predisposition Study); CDISC (Clinical Data Interchange Standards Consortium); CDISC-GLOSS (CDISC Glossary Terminology); Cellosaurus; CPTAC (NCI’s Clinical Proteomic Tumor Analysis Consortium); CRCH (Cancer Research Center of Hawaii Nutrition Terminology); CTCAE (Common Terminology Criteria for Adverse Events); CTDC (NCI’s Clinical Trial Data Commons); CTEP (Cancer Therapy Evaluation Program); CTRP (Clinical Trials Reporting Program); DIPG/DMG (Diffuse Intrinsic Pontine Glioma/Diffuse Midline Glioma Research Network); EDQM-HC (European Directorate for the Quality of Medicines & Healthcare); FDA (U.S. Food and Drug Administration); GAIA (Global Alignment of Immunization safety Assessment in pregnancy Terminology); GDC (NCI;s Genomic Data Commons); HL7 (Health Level 7); ICD-10 (International Classification of Diseases, Tenth Revision); ICDC (NCI’s Integrated Canine Data Commons); ICH (International Conference on Harmonization); INC (International Neonatal Consortium – Critical Path Institute); MedDRA (Medical Dictionary for Regulatory Activities); NAACCR (North American Association of Central Cancer Registries); NCCN (National Comprehensive Cancer Network); NCD (FDA’s National Drug Code); NCPDP (National Council for Prescription Drug Programs); OORO (Operational Ontology for Radiation Oncology); ORCHESTRA (Multinational SARS-Cov2 Project); PCDC (Pancreatic Cancer Detection Consortium); PI-RADS (Prostate Imaging-Reporting and Data System); SEER (NCI’s Surveillance, Epidemiology, and End Results); SeroNet (NCI’s Serological Sciences Network); UCUM (Unified Code for Units of Measure)].
Each individual concept in NCIt is assigned a unique, permanent NCI C-code (C stands for CUI, or Concept Unique Identifier), which ensures uniqueness in those instances when a preferred name or synonym may be identical to another concept but whose semantic meaning is distinct; it is also given a definition, which ensures that humans and systems have a common understanding of the concept such that the concept is used accurately, precisely, and consistently across multiple users and systems. While some un-defined concepts still exist, the vast majority of concepts in NCIt have an assigned NCI definition. Contributing concept source owners can choose to tag and maintain their own definitions (using the ALT_DEFINITION property) as long as the meaning directly corresponds to the NCI definition. Definitional differences between sources may be as a result of source definition writing rules, to ensure consistency across definitions of terms of the same type, or the difference in audience for which the definitions will be used, such as the difference between lay and technical language.
NCIt term types, properties, and relationships allow the editor to assign multiple terms types to a single, unique concept. This ensures the clear identification of a concept’s one to many term types (eg, preferred terms, synonyms, abbreviations, adjectival forms, antiquated names), which are clearly delineated by term source. NCIt structural elements also allow for the bundling of concepts into value sets using the Concept_in_Subset property. Indeed, NCI-EVS currently maintains over 1500 value sets for 26 organizational entities including the US FDA, CDISC, the European Directorate for the Quality of Medicines & Healthcare (EDQM), NCI programs such as Clinical Trials Reporting Program (CTRP) and Surveillance, Epidemiology, and End Results (SEER), and the National Council for Prescription Drug Programs (NCPDP).18 This subset bundling functionality allows source contributors to control the development and maintenance of external model-dependent value sets without relying on NCIt to build artificial hierarchies to support those external models.
NCI Metathesaurus (NCIm)
The NCI Metathesaurus (NCIm) is a free, non-license restricted biomedical terminology database that provides rich synonymy and mappings among codes and terms in commonly used biomedical terminologies. NCIm cross-links the content from NCIt to a wide variety of terminologies and ontologies that are published separately, such as Common Terminology Criteria for Adverse Events (CTCAE), Gene Ontology (GO), the International Classification of Diseases (ICDs), the Logical Observation Identifier Names and Codes (LOINC), the Medical Dictionary for Regulatory Activities (MedDRA), and the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT). Altogether, the database contains 8 million terms (preferred terms, synonyms, abbreviations, etc.) mapped to over 3.5 million concepts from 100 source terminologies, with over 24 million modeled relationships. For example, the concept of ‘Heart’ is mapped to 27 terms across 17 source vocabularies within the NCIt.19 Some proprietary vocabularies are also included and have restrictions on their use.20
NCIm content is accessible through a public browser (ncim.nci.nih.gov) and the LexEVS API. Many NCIm structural elements are similar to those found in NCIt, including source-tagged definitions and term types. In addition, the term code field contains the source code identifier from the original source terminology, which can assist in mapping activities when translating from one biomedical coding system to another. Vocabulary insertion into NCIm follows a well-established editing process including both human and machine processing.21
Standalone Vocabularies and Terminology Mappings
EVS makes a number of external vocabularies and ontologies easily accessible on the NCI Term Browser platform.22 These vocabularies generally reflect the NIH community interests. NCI-EVS also generates and provides pairwise mappings between several of its hosted vocabularies to support data translation and cross-referencing. Likewise, these mappings generally reflect NIH community interests. Each mapping contains a pairwise, synonymous linkage between the source code and target code of the hosted vocabularies. These mappings are viewable and browsable in the NCI Term Browser environment and are also CSV exportable. Notable mappings include NCIt to MedDRA and ICD-03, SNOMEDCT_US to ICD-10, and NCIt to SwissProt (Swiss Protein Sequence Database), among others.
EVS and CDISC Collaboration: CDISC Controlled Terminology
History
The NCI-EVS-CDISC collaboration began nearly 20 years ago, when CDISC embarked on identifying a terminology systems provider to support its data standards development activities. NCI-EVS was chosen to partner with CDISC because of its freely and easily accessible terminology browsers and publication products, robust semantics associated with each stored concept, established terminology infrastructure and technology services, and its ability for individual source tagging and control of semantic content within the wider ecosystem of NCIt. Additionally, NCI-EVS was already established as the terminology services provider for the US FDA.10,23 This multi-year collaboration has produced a robust terminology development process and a large array of published CDISC terminology products, ensuring the CDISC terminology program’s continued success in supporting the development and maintenance of CDISC standards.
Terminology Standards Development Process
CDISC terminology development adheres to CDISC Operating Procedure COP-001,24 which specifies the terminology standards development process by and for the CDISC user community and including public review and comment on all content. The CDISC user community interacts directly with the CDISC terminology teams, which are themselves made up of individuals from the CDISC community, through the CDISC Term Suggestion form.25 Types of valid requests include the addition of terms to existing codelists, the addition of a new codelist(s), modifications to published terms (e.g., synonym or definition changes), new (or modifications to existing) codetable mapping files, new (or modifications to existing) CDISC CT team rules documents, and terminology implementation questions. NCI-EVS maintains the CDISC Term Suggestion form and NCI-EVS personnel interact directly with requesters to acknowledge receipt of the request and to all follow-up communications with requesters on behalf of the CDISC CT teams.
Requests that come through the CDISC Term Suggestion form are integrated into CDISC terminology team working documents on a weekly basis. NCI-EVS personnel do background research on each request, manage the weekly agendas for each team, and lead or co-lead all CDISC CT teams. CDISC has a number of active terminology teams (20 teams at the time of publication) that develop terminology based on subject matter areas related to CDISC standards. These volunteer teams are composed of clinical and non-clinical subject matter experts (SMEs), terminologists, data modelers, and data standards experts from pharmaceutical, biologics, and device companies; contract research organizations (CROs); academic institutions; and global regulators. Within each team, term requests are generally resolved in the order they are received.
The products of this collaborative effort, including new terms (CDISC Submission Values, Synonyms, and Definitions) and codelists, changes to existing terms, and denied request reasoning, are bundled together at the end of each quarter and made available to the public for review and comment. This quarterly CDISC CT public review is open to all and is administered through CDISC’s Jira issue tracker. CDISC CT public review runs for four weeks followed by comment resolution and terminology updates. The CDISC CT package then undergoes additional NCI-EVS personnel-led content quality control (QC) and processing prior to loading into Protégé. An additional QC step is performed after database processing to ensure all new terms and changes are complete in Protégé prior to terminology publication.
CDISC publication files are generated through Protégé reporting tools at the end of each quarter. CDISC CT is presented in the following file formats; .txt, .xls, .pdf, .html, OWL/RDF, and odm.xml. Diff files are also generated that compare the previous quarter’s terminology to the most recent version. This program was developed by NCI-EVS personnel and is available for general use on a GitHub site.26 CDISC CT is versioned using a date identifier that corresponds to the publication date. CDISC Terminology publication files, diff files, and ReadMe documents are stored in the CDISC folder of the NCI Ftp (File Transfer Protocol) server.27 Each sub-folder contains an Archive folder in which all previous versions of CDISC CT and supplemental files are stored indefinitely. CDISC maintains a CDISC CT Publication Schedule on its website with planned quarterly publication dates up to a year in advance.28
CDISC Controlled Terminology in NCIt
The NCI-EVS-CDISC collaboration has yielded a large, mature set of carefully curated terminology standards. Just as CDISC standards have expanded and diversified over the years, so too have CDISC terminology holdings within NCIt. As of the end of December 2022, there are 27 115 concepts tagged as CDISC terms in NCIt, which contain nearly 55 000 CDISC-tagged submission values. All CDISC-tagged concepts contain a CDISC-sourced ALT_Definition to ensure that all users understand and use the concept in the same way. NCI-EVS maintains and publishes eight subsets of CDISC terminology, each with one-to-many codelists (Table 1). As of the end of December 2022, there were over 1 300 published codelists associated with CDISC terminology in NCIt.
Published CDISC subsets and codelists as of end December 2022. [ADaM (Analysis Data Model); DDF (Digital DataFlow); CDASH (Clinical Data Acquisition Standards Harmonization); SEND (Standard for the Exchange of Nonclinical Data); SDTM (Study Data Tabulation Model)].
| Terminology Subset | Codelist Numbers |
|---|---|
| ADaM Terminology | 14 |
| Define-XML Terminology | 15 |
| DDF Terminology | 39 |
| CDASH Terminology | 22 |
| CDISC Glossary | 1 |
| Protocol Terminology | 47 |
| SEND Terminology | 125 |
| SDTM Terminology | 1 045 |
| Total | 1 308 |
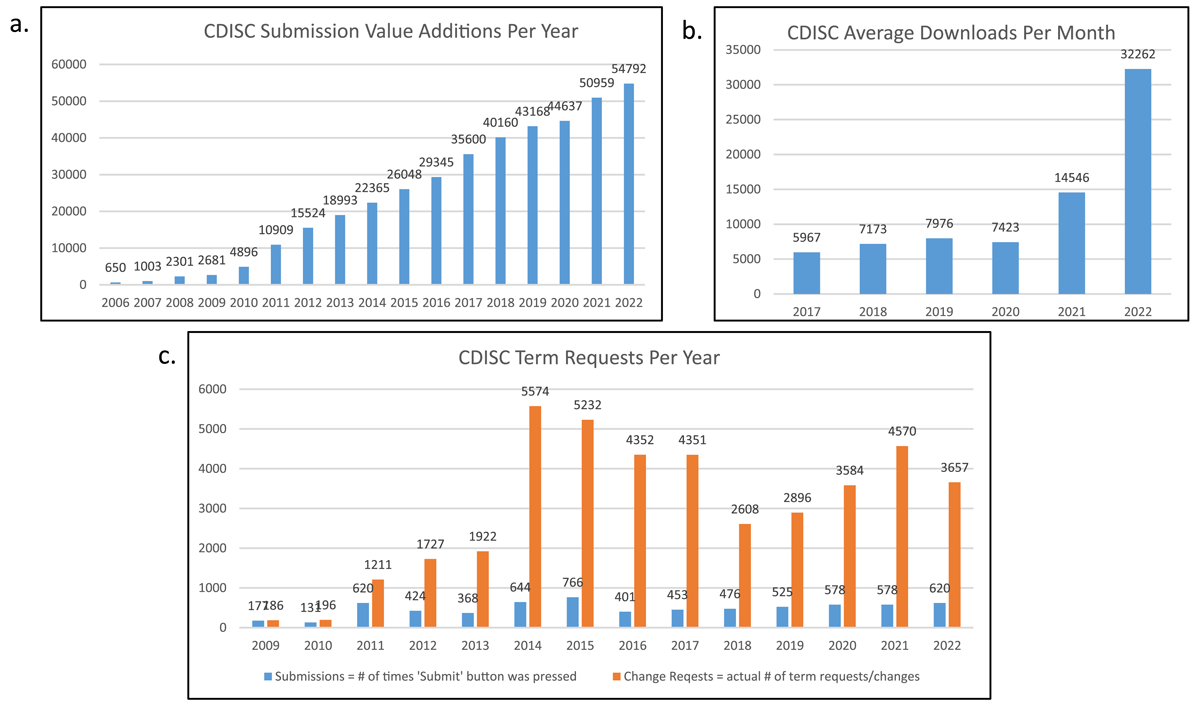
The CDISC terminology set has seen significant growth since 2006 (Figure 2a), growing from an initial set of 650 terms to nearly 55 000 as of the end of December 2022. Download numbers are also increasing year over year, with more than 32 000 average downloads per month in 2022 (Figure 2b). This growth in terminology is driven by foundational standards, therapeutic area (TA) standards, and CDISC user requests through the CDISC Term Request form (Figure 2c).
Integration of CDISC terminology within the NCIt infrastructure enables free and simple accessibility, and harmonization and mapping across multiple sources and other standards. The advantages of integration include contributing source content control, effective knowledge representation of semantically complex domains via concept definitions, ontological relationships and enrichment with synonyms, value set maintenance in support of data models, meticulously scheduled terminology publications, and access to NCI-EVS subject matter expertise and technical resources. In turn, the CDISC and NCI-EVS joint effort meets the NCI’s strategic goals and initiatives to support, through collaborations with standards development organizations (SDO), the development of interoperability standards; and to promote meaningful exchange, ‘FAIRness’ (Find, Access, Interoperate, and Reuse), and the sharing of cancer research data.
Supplemental Terminology Products from NCI-EVS and CDISC
Beyond this large set of CDISC terminology standards, NCI-EVS and CDISC create and maintain supplemental terminology products that are available on the NCI Ftp server and the CDISC website. These supplemental terminology files aid the CDISC user community in the management of the terminology versions, in conforming collected data to CDISC submission values, in CRF design, in the creation of value level metadata, and in explicitly defining relationships between and among CDISC terminology and model elements, all of which are described in Table 2.
CDISC supplemental terminology products and their descriptions.
| Supplemental CT Product Name | Supplemental CT Product Description |
|---|---|
| CT Relationships28 | Stored on the CDISC website – Relationships exist between CDISC codelists and CDISC model variables. As a result of differences in publication frequency, a late binding effect may be created between CDISC data models, implementation guides, therapeutic area (TA) user guides, and quarterly CDISC CT publications. This product provides explicit and complete machine and human readable linkages between CDISC CT and CDISC model elements across all CDISC standards documents. |
| Terminology diff files29 | Stored on NCI’s Cancer.gov website – Each quarterly CDISC terminology data release includes files that contain programmatically generated changes from the previous quarter’s release. Comparison of changes over different time periods for purposes of terminology analysis or mapping can be done by individuals using the free source code for the Java-based program developed by NCI-EVS to create such files.30 |
| Unit-UCUM Mapping file31 | Stored on the CDISC website – To conform to Unified Code for Units of Measure (UCUM) units of measure and CDISC submission values, a mapping file is maintained between published CDISC unit concepts and one-to-many synonymous UCUM expressions. This facilitates the mapping of a collected UCUM-compliant unit of measure to the appropriate CDISC submission value. |
| Codetable Mapping files32 | Stored on the CDISC website – The large number of relationships between and among published terms in CDISC CT are not present in the terminology publication files. Codetable Mapping files provide machine and human readable relationships between published terms within multiple codelists across a single domain context. This file can be used for CRF building, QA/QC, and data mapping. |
| LOINC-LB Mapping file33 | Stored on the CDISC website – The US FDA requires the submission of Logical Observation Identifiers Names and Codes (LOINC®) within clinical LB (Laboratory Findings) domain datasets under certain circumstances.34 This mapping file contains examples of how the individual parts of a LOINC code map to CDISC LB domain variables and CT, which may assist lab and instrument vendors in providing the correct data for CDISC standardization. |
Notable CDISC Terminology Projects and Value Sets
Supporting a family of standards, such as the CDISC standards requires multiple terminology projects supporting distinct terminology products. Some examples include the CDISC Therapeutic Area Standards, the SEND-INHAND collaboration, and the CDISC QRS standards.
CDISC Therapeutic Area Standards
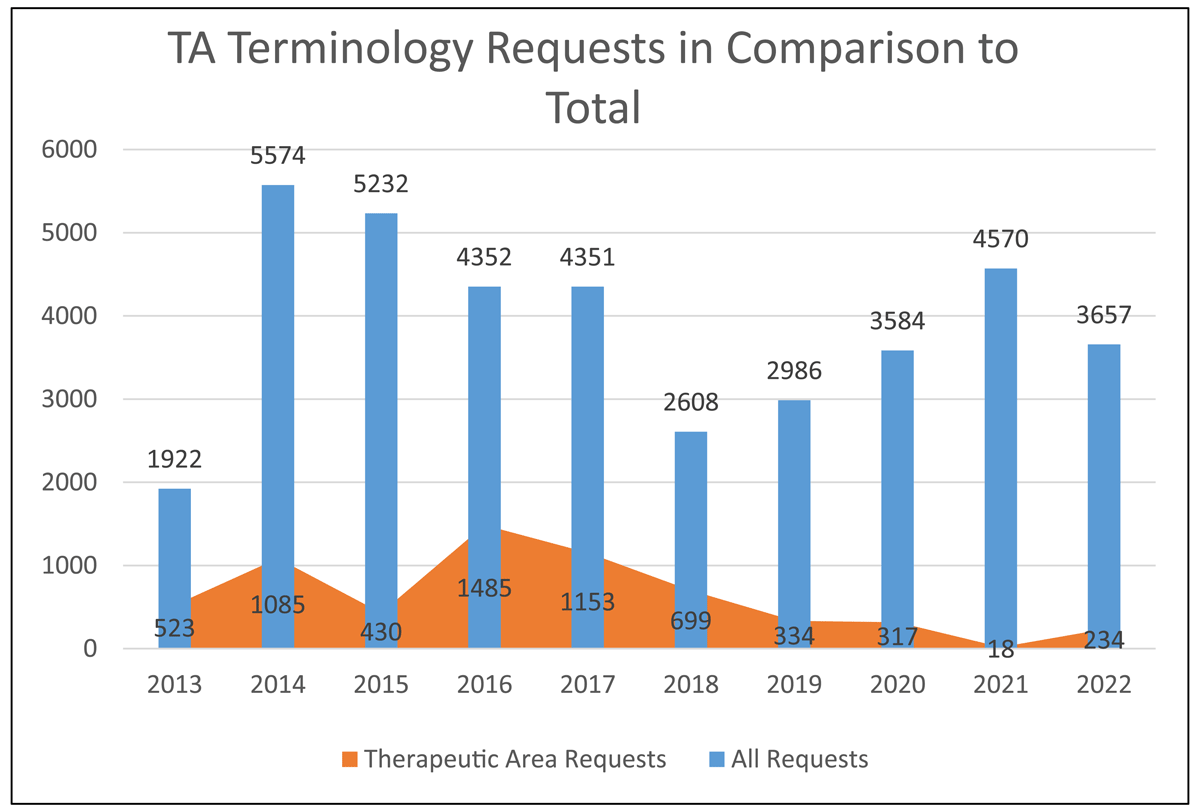
The CDISC therapeutic area (TA) standards development program began in 2012, with the purpose of extending the foundational standards to cover therapeutic-area specific data and endpoints. The published therapeutic area user guides (TAUGs) contain disease-specific metadata, implementation advice, CDASH-compliant CRFs (with example valid value lists), example SDTM datasets (with example CDISC CT), TA-specific non-standard variables, and example analysis datasets. To date, 49 TAUGs have been published or are currently in development that span autoimmune, cardiovascular, endocrine, gastrointestinal, infectious, mental health, neurological, oncological, rare, respiratory and other diseases and disorders.35,36,37 To date, about 6 000 term requests have been submitted by TA teams to support the TAUGs, though the vast majority of CDISC terminology requests are made by the user community (Figure 3). New terminology development is a large part of the TAUG development process, whether the teams are extending existing codelists or creating new codelists to support new variables and domains. Draft terminology analysis may also lead to model refinement as feedback recommendations from CDISC CT teams are fed back to the TAUG development team. For example, terminology analysis of new test values in a particular findings domain sometimes conclude with the recommendation that a different domain and data structure be used to model the data. Additionally, inconsistent modeling strategies for similar data may be identified early, owing to the terminology team’s familiarity with term requests from previous TAUG development teams. This feedback loop between terminologists, SMEs, and data modelers ensures high quality and consistent data standards.
Oncology TA Standards – To date, CDISC has published five oncology-related TAUGs, including the Breast Cancer Therapeutic Area User Guide v1.0, the Colorectal Cancer Therapeutic Area User Guide v1.0, the Pancreatic Cancer Therapeutic Area User Guide v1.0, the Prostate Cancer Therapeutic Area User Guide v1.0, and the Lung Cancer Therapeutic Area User Guide v1.0.38,39,40,41,42. As part of these development efforts, NCI-EVS has created and maintains SDTM Terminology containing terms to support 84 tumor response criteria, including multiple versions of the RECIST tumor response criteria with representative value level metadata in published oncology Codetable Mapping files. Additionally, terminology to support the AJCC v7 TNM Staging system has been incorporated into CDISC standards as a clinical classification.
COVID-19 TA Standards –The global COVID-19 pandemic brought into sharp relief the immediate need for data standards to support the development of SARS-CoV2-related therapies and vaccines, as well as data standards pertaining to pandemic-related changes to ongoing trials. In May 2020, CDISC published the provisional standards: The COVID-19 Therapeutic Area User Guide v1.0; Guidance for Ongoing Studies Disrupted by COVID-19; and Vaccine Administration v1.0.43 The development timeline of these therapeutic area user guides necessitated an additional, out of cycle CDISC CT terminology publication in May 2020.
SEND-INHAND Terminology Collaboration
NCI-EVS develops, publishes, and maintains the controlled terminology to support the CDISC Standard for the Exchange of Non-Clinical Data (SEND) data model and implementation guides.44 Based on US FDA requirements, the SEND pathology terminology standards are harmonized with the Society of Toxicologic Pathology’s (STP) International Harmonization of Nomenclature and Diagnostic Criteria for Lesions in Rats and Mice (INHAND) standard nomenclature for proliferative and non-proliferative findings in rodent and non-rodent species.45 The INHAND nomenclature is available in pdf publications on the global open Registry Nomenclature Information System (goRENI) platform.46
The SEND CT team, in coordination with INHAND organ working groups, has harmonized the concepts in the CDISC Neoplasm Type (NEOPLASM) and Non-Neoplastic Finding Type (NONNEO) codelists, which support the MISTRESC variable in the SEND MI (Microscopic Findings) domain, to INHAND terminology.47 The CDISC submission values within these value sets represent the INHAND concept, if available, but may not be identical due to SEND Terminology business rules. These codelists may also contain terminology beyond the scope of INHAND terms that are in use by the CDISC user community. Likewise, the CDISC definitions reflect the usage of the concepts in the INHAND nomenclature and, in many cases, cite INHAND descriptions directly. This marriage of nomenclature to systems-implementable controlled terminology formats positively promotes the use of standard nomenclature in regulated non-clinical data submissions. Through harmonization activities such as these, the use of CDISC controlled terminology for data domains has been shown to positively promote cross-study analysis through semantic interoperability.48 Further, harmonization such as between basic science, pre-clinical studies, and clinical studies builds semantic bridges that can facilitate drug discovery and drug repurposing, through the explicit definition and harmonization of common biologic, physiologic, metabolic, and pharmacologic concepts and relationships.
CDISC QRS Standards
Patient input and outcome measures are an integral part of the US FDA’s Drug Development Tools Qualification Program for clinical outcome assessment (COA) instruments, as described by PDUFA VI.49 The US FDA classifies COAs into four types: clinician-reported outcome (ClinRO), observer-reported outcome (ObsRO), patient-reported outcome (PRO) and performance outcome (PerfO).50 CDISC Questionnaires, Ratings and Scales (QRS) controlled terminology and data standards supplements assist in structuring all four types of COA data to ensure the use of standardized formats in data collection and reporting.
Currently, NCI-EVS and CDISC develop controlled terminology and supplements for three types of instruments: Questionnaires, Functional Tests, and Clinical Classifications, which are stored in the CDISC QS, FT and RS domains, respectively. Information about the definitions, classifications and SDTM domains required to model and store data pertaining to the three types of instruments are described elsewhere.51
The development process for CDISC QRS controlled terminology follows the CDISC terminology development process but includes an additional copyright/public domain verification step and, for copyrighted instruments, it includes an official request to the copyright holder for permission to develop data standards and terminology.51,52,53 Additionally, as digital biomarkers are developed and validated, the QRS terminology will continue to grow to include new data types. As of the 2022-12-16 publication of CDISC SDTM terminology, NCI-EVS and CDISC QRS CT teams have developed 20 270 QRS test, category, and response terms. Additionally, CDISC has published more than 200 instrument-specific QRS supplements to date.
The Use of CDISC Terminology in RWD/RWE
The robust technology, services, and processes that NCI-EVS employs to support the CDISC terminology program has yielded a terminology set that is robust, fit for purpose, concisely defined, and ensures semantic interoperability for efficient regulatory review of medical products. These same technologies, services and processes will support the current effort to expand the use of CDISC standards beyond prospective data collection for clinical and non-clinical research, particularly with regards to the application of CDISC data standards for RWD, and RWE generation. Refer to the published CDISC Glossary for standard definitions of RWD and RWE.54
RWE, which is generated from RWD, has many applications that include postmarketing safety surveillance, pharmacovigilance, adverse event reporting, the development of guidelines and decision support tools for clinicians, insurance plan payer decision making, and complementing findings from randomized clinical trials (RCTs). Global regulators are increasingly looking at how RWD and RWE can enhance regulated clinical research.13,55,56,57 However, disparate data sources, data models, terminologies, and data exchange standards that currently support RWD collection and analysis are not interoperable. This currently necessitates manual mapping between RWD and clinical trial data, which invites loss of information (detail) by mapping more granular data to higher level concepts in common between the RWD and research data. The traceability of these transformations isn’t always rigorous enough to bolster regulatory decisions.58 Harmonizing CDISC standards and terminology with those used in clinical RWD and promoting rigorous data standardization in RWD areas where none exists may provide the key to unlocking the promise of RWD and RWE for regulatory applications.59,60,61
Recently, the US FDA published draft Guidance document that detailed how RWD and RWE could be integrated into clinical research data.62,63 Currently, sponsors submitting clinical and non-clinical study data derived from real world sources should use those data standards and terminologies listed in the US FDA’s Data Standards Catalog and it is suggested that a conforming (or mapping) step should be undertaken to convert terminology values from RWD sources to those terminologies used in clinical research, including CDISC terminology.10,64 The terminology mappings provided by NCI-EVS between commonly used biomedical coding dictionaries, including those used in the healthcare space, may provide a more rigorous method for to this conformance step, and provide the required traceability. The NCI Metathesaurus may also be a useful tool in this effort. Mappings are costly to generate and maintain and there is concern about information loss, i.e., if mapped or conformed, do values truly represent the source data from which they are obtained?65,66,67 More investigation will be required to understand how these terminology mappings affect data integrity.
Conclusions
CDISC terminology underpins and supports CDISC’s foundational and therapeutic area standards. NCI-EVS has ensured the robustness and quality of these standards through the development, publication, and maintenance of CDISC Controlled Terminologies in the NCI Thesaurus. NCI-EVS has assured the successful growth of the CDISC Terminology program through its established semantic infrastructure and terminology development and management expertise. Lessons learned and process iteration have ensured that the CDISC terminology development program has been able to support the growing complexity of the CDISC standards to meet the needs of international regulators and researchers. NCI-EVS enables semantic interoperability and data integration across multiple data standards and models through its robust terminology platforms, NCIt and NCIm. Harmonization activities between nomenclature and systems-implementable controlled terminology formats positively promotes cross-study analysis in both the pre-clinical and clinical space. The cross-terminology mappings provided by NCI-EVS between commonly used biomedical coding dictionaries, including those used in the healthcare space, may provide a more rigorous method for mapping/conforming data from disparate sources, as well as providing the required traceability. As CDISC looks ahead to applications of its data standards in settings outside of clinical and non-clinical research, the good terminology practices, definitions, and semantic infrastructure provided by NCI-EVS will provide more rigor to RWD (whether through direct use or conforming), to better support the generation of RWE.
Competing Interests
[[COMPETING INTEREST STATEMENT TO BE PROVIDED]]
References
1. National Institutes of Health. National Cancer Institute EVs wiki. Accessed January 31, 2023. https://wiki.nci.nih.gov/display/EVS.
2. National Institutes of Health. NCI thesaurus. Accessed January 31, 2023. https://ncit.nci.nih.gov/ncitbrowser
3. National Institutes of Health. NCI Metathesaurus. Accessed January 31, 2023. https://ncim.nci.nih.gov/ncimbrowser/
4. Fragoso G, de Coronado S, Haber M, Hartel F, Wright L. Overview and utilization of the NCI Thesaurus. Comp Funct Genomics. 2004; 5(8): 648–654. DOI: http://doi.org/10.1002/cfg.445
5. de Coronado S, Wright LW, Fragoso G, et al. The NCI Thesaurus quality assurance life cycle. J Biomed Inform. 2009; 42(3): 530–539. DOI: http://doi.org/10.1016/j.jbi.2009.01.003
6. US Food and Drug Administration. Providing Regulatory Submissions In Electronic Format — Standardized Study Data Guidance for Industry. Published June 2021. Accessed April 2023. https://www.fda.gov/media/82716/download
7. Japan Pharmaceutical and Medical Devices Agency. Utilization of real world data – PMDA’s approaches. Published March 23, 2021. Accessed January 2023. https://www.pmda.go.jp/english/about-pmda/0004.pdf
8. China National Medical Products Administration. Guideline on the submission of clinical trial data. Published Oct 1, 2020. Accessed January 2023. https://www.pharmews.xyz/2020/05/guideline-on-submission-of-clinical.html; https://www.cde.org.cn/main/news/viewInfoCommon/d238c109af91307fb4f16c4da86506b6
9. European Medicines Agency. HMA-EMA Joint Big Data Taskforce – summary report. Published March 2017. Accessed January 2023. https://www.ema.europa.eu/en/documents/minutes/hma/ema-joint-task-force-big-data-summary-report_en.pdf
10. US Food and Drug Administration. Study Data Standards Resources. Published May 2022. Accessed January 2023. https://www.fda.gov/industry/fda-data-standards-advisory-board/study-data-standards-resources
11. Japanese Pharmaceuticals and Medical Devices Agency. New Drug Review with Electronic Data website. Accessed January 2023. https://www.pmda.go.jp/english/review-services/reviews/0002.html
12. US Food and Drug Administration. Framework for FDA’s Real-World Evidence Program. Published December 2018. Accessed January 2023. https://www.fda.gov/media/120060/download
13. Eichler HG, Pignatti F, Schwarzer-Daum B, et al. Randomized Controlled Trials Versus Real World Evidence: Neither Magic Nor Myth. Clin Pharmacol Ther. 2021; 109(5): 1212–1218. DOI: http://doi.org/10.1002/cpt.2083
14. International Organization for Standardization. ISO/IEC 11179-4:2004 Information technology — Metadata registries (MDR) — Part 4: Formulation of data definitions. Second edition 2004-07-15.
15. Cimino JJ, Hayamizu TF, Bodenreider O, Davis B, Stafford GA, Ringwald M. The caBIG terminology review process. J Biomed Inform. 2009; 42(3): 571–580. DOI: http://doi.org/10.1016/j.jbi.2008.12.003
16. National Institutes of Health. Term suggestion. Accessed January 2023. https://ncitermform.nci.nih.gov/ncitermform/
17. Protégé. Accessed January 2023. https://protege.stanford.edu/
18. National Institutes of Health. NCI term browser – value set source view. https://ncit.nci.nih.gov/ncitbrowser/ajax?action=create_src_vs_tree&nav_type=valuesets&mode=0. Accessed January 2023.
19. National Cancer Institute. NCI Metathaurus. Heart (CUI C0018787). Accessed March 2023. https://ncim.nci.nih.gov/ncimbrowser/ConceptReport.jsp?dictionary=NCI%20Metathesaurus&code=C0018787&type=synonym&sortBy=source#SynonymsDetails
20. National Institutes of Health. NCI Metathesaurus. NCI wiki. https://wiki.nci.nih.gov/pages/viewpage.action?pageId=8160732. Accessed January 31, 2023.
21. National Institutes of Health. Accessed January 2023. https://wiki.nci.nih.gov/display/EVS/Metathesaurus+Editing%3A+NCI-NLM+Editing+Systems+Workgroup
22. National Institutes of Health. NCI term browser. Accessed January 2023. https://ncit.nci.nih.gov/ncitbrowser/pages/multiple_search.jsf?nav_type=terminologies
23. National Cancer Institute Center for Biomedical Informatics and Information Technology. FDA terminology. https://datascience.cancer.gov/resources/cancer-vocabulary/fda-terminology. Accessed January 2023.
24. Clinical Data Interchange Standards Consortium. CDISC Operating Procedure COP-001 Standards Development. Accessed January 2023. https://www.cdisc.org/system/files/about/cop/CDISC-COP-001-Standards_Development_2019.pdf
25. National Institutes of Health. Term suggestion. Accessed January 31, 2023. https://ncitermform.nci.nih.gov/ncitermform/?version=cdisc
26. National Institutes of Health. NCI-EVS. NCI-Diff-CDISC GitHub. Accessed March 2023. https://github.com/NCIEVS/nci-diff-cdisc)
27. National Institutes of Health. Index of /FTP1/CDISC. Accessed January 2023. https://evs.nci.nih.gov/ftp1/CDISC/
28. Clinical Data Interchange Standards Consortium. Controlled terminology. Accessed January 2023. https://www.cdisc.org/standards/terminology/controlled-terminology#standard__Resources
29. National Cancer Institute Center for Biomedical Informatics and Information Technology. CDISC terminology. Accessed January 2023. https://datascience.cancer.gov/resources/cancer-vocabulary/cdisc-terminology
30. GitHub. NCIEVS/NCI-diff-CDISC: Diff specific for CDISC reports. Accessed January 2023. https://github.com/NCIEVS/nci-diff-cdisc.
31. Clinical Data Interchange Standards Consortium. Unit-UCUM Mapping File. Accessed January 2023. https://www.cdisc.org/standards/terminology/controlled-terminology#standard__Unit-UCUM_Mapping_File.
32. Clinical Data Interchange Standards Consortium. Codetable Mapping Files. Accessed January 2023. https://www.cdisc.org/standards/terminology/controlled-terminology#standard__Codetable_Mapping_Files
33. Clinical Data Interchange Standards Consortium. LOINC to LB Mapping Files. Accessed January 2023. https://www.cdisc.org/standards/terminology/controlled-terminology#standard__LOINC_to_LB_Mapping_Files.
34. US Food and Drug Administration. Recommendations for the Submission of LOINC® Coides in Regulatory Applications to the US Food & Drug Adminiations. Guidance for Industry November 2017. Accessed January 2023. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/recommendations-submission-loincr-codes-regulatory-applications-us-food-and-drug-administration
35. Mullin AP, Corey D, Turner EC, et al. Standardized Data Structures in Rare Diseases: CDISC User Guides for Duchenne Muscular Dystrophy and Huntington’s Disease. Clin Transl Sci. 2021; 14(1): 214–221. DOI: http://doi.org/10.1111/cts.12845
36. Neville J, Kopko S, Romero K, et al. Accelerating drug development for Alzheimer’s disease through the use of data standards. Alzheimers Dement (N Y). 2017; 3(2): 273–283. Published 2017 Apr 15. DOI: http://doi.org/10.1016/j.trci.2017.03.006
37. Perrone RD, Neville J, Chapman AB, et al. Therapeutic Area Data Standards for Autosomal Dominant Polycystic Kidney Disease: A Report From the Polycystic Kidney Disease Outcomes Consortium (PKDOC). Am J Kidney Dis. 2015; 66(4): 583–590. DOI: http://doi.org/10.1053/j.ajkd.2015.04.044
38. Clinical Data Interchange Standards Consortium. Therapeutic Area User Guide for Breast Cancer Version 1.0 (Provisional) (Released 2016-05-16). Accessed January 2023. https://www.cdisc.org/standards/therapeutic-areas/breast-cancer
39. Clinical Data Interchange Standards Consortium. Colorectal Cancer Therapeutic Area User Guide Version 1.0 (Released 2018-11-15). Accessed January 2023. https://www.cdisc.org/standards/therapeutic-areas/colorectal-cancer
40. Clinical Data Interchange Standards Consortium. Therapeutic Area User Guide for Pancreatic Cancer Version 1.0 (Provisional) (Released 2021-10-05). Accessed January 2023. https://www.cdisc.org/standards/therapeutic-areas/pancreatic-cancer
41. Clinical Data Interchange Standards Consortium. Therapeutic Area User Guide for Prostate Cancer Version 1.0 (Provisional) (Released 2017-07-10). Accessed January 2023. https://www.cdisc.org/standards/therapeutic-areas/prostate-cancer.
42. Clinical Data Interchange Standards Consortium. Therapeutic Area Data Standards User Guide for Lung Cancer Version 1.0 (Provisional) (Released 2019-05-06). Accessed January 2023. https://www.cdisc.org/standards/therapeutic-areas/lung-cancer.
43. Clinical Data Interchange Standards Consortium. Therapeutic Area User Guide for COVID-19 Version 1.0 (Provisional) (Released 2021-07-08). Accessed January 2023. https://www.cdisc.org/standards/therapeutic-areas/covid-19
44. Clinical Data Interchange Standards Consortium. Standard for the Exchange of Non-Clinical Data website. Accessed January 2023. https://www.cdisc.org/standards/foundational/send
45. Keenan CM, Goodman DG. Regulatory Forum commentary: through the looking glass—SENDing the pathology data we have INHAND. Toxicol Pathol. 2014; 42(5): 807–810. DOI: http://doi.org/10.1177/0192623313485451
46. Global open RENI. GoRENI Version 3.19.34. Accessed January 2023. https://www.goreni.org/
47. Keenan CM, Baker J, Bradley A, et al. International Harmonization of Nomenclature and Diagnostic Criteria (INHAND): Progress to Date and Future Plans. Toxicol Pathol. 2015; 43(5): 730–732. DOI: http://doi.org/10.1177/0192623314560031
48. Carfagna MA, Bjerregaard TG, Fukushima T, et al. SEND harmonization & cross-study analysis: A proposal to better harvest the value from SEND data. Regul Toxicol Pharmacol. 2020; 111: 104542. DOI: http://doi.org/10.1016/j.yrtph.2019.104542
49. US Food and Drug Administration, Center for Drug Evaluation and Research. PDUFA VI: Fiscal years 2018 – 2022. Accessed January 2023. https://www.fda.gov/industry/prescription-drug-user-fee-amendments/pdufa-vi-fiscal-years-2018-2022.
50. US Food and Drug Administration, Center for Drug Evaluation and Research. Clinical outcome assessment (coa):faqs. Accessed January 2023. https://www.fda.gov/about-fda/clinical-outcome-assessment-coa-frequently-asked-questions
51. Clinical Data Interchange Standards Consortium. QRS. Accessed January 31, 2023. https://www.cdisc.org/standards/foundational/qrs.
52. QRS Addendum to the CDISC Operating Procedure COP-001 Standards Development. CDISC. https://www.cdisc.org/system/files/about/cop/CDISC-COP-001-Standards_Development_2019.pdf. Accessed January 2023.
53. Clinical Data Interchange Standards Consortium. QRS Naming Rules. Accessed January 2023. https://www.cdisc.org/sites/default/files/2021-03/QRS_Naming_Rules_2021-03-05.xlsx.
54. National Institutes of Health. CDISC Glossary, 2021-12-25. NCI-EVS Terminology Resources. Accessed January 2023. https://evs.nci.nih.gov/ftp1/CDISC/Glossary/
55. US Food and Drug Administration. 21st Century Cures Act website. Published January 2020. Accessed January 2023. https://www.fda.gov/regulatory-information/selected-amendments-fdc-act/21st-century-cures-act
56. US Food and Drug Administration. Framework for FDA’s Real World Evidence Programme. December 2018. Accessed March 2023. https://www.fda.gov/media/120060/download
57. Ahn EK. A brief introduction to research based on real-world evidence: Considering the Korean National Health Insurance Service database. Integr Med Res. 2022; 11(2): 100797. DOI: http://doi.org/10.1016/j.imr.2021.100797
58. Collins R, Bowman L, Landray M, Peto R. The Magic of Randomization versus the Myth of Real-World Evidence. N Engl J Med. 2020; 382(7): 674–678. DOI: http://doi.org/10.1056/NEJMsb1901642
59. Collins R, Bowman L, Landray M, Peto R. The Magic of Randomization versus the Myth of Real-World Evidence. N Engl J Med. 2020; 382(7): 674–678. DOI: http://doi.org/10.1056/NEJMsb1901642
60. Facile R, Muhlbradt EE, Gong M, et al. Use of Clinical Data Interchange Standards Consortium (CDISC) Standards for Real-world Data: Expert Perspectives From a Qualitative Delphi Survey. JMIR Med Inform. 2022; 10(1): e30363. Published 2022 Jan 27. DOI: http://doi.org/10.2196/30363
61. Biedermann P, Ong R, Davydov A, et al. Standardizing registry data to the OMOP Common Data Model: experience from three pulmonary hypertension databases. BMC Med Res Methodol. 2021; 21(1): 238. Published 2021 Nov 2. DOI: http://doi.org/10.1186/s12874-021-01434-3
62. US Food and Drug Administration. Real-world data (RWD) and real-world evidence (RWE) are playing an increasing role in health care decisions. Real-World Evidence website. Accessed January 2023. https://www.fda.gov/science-research/science-and-research-special-topics/real-world-evidence.
63. US Food and Drug Administration. Considerations for the Use of Real-World Data and Real-World Evidence to Support Regulatory Decision-Making for Drug and Biological Products. Draft Guidance for Industry 12/07/2021. Accessed January 2023. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/considerations-use-real-world-data-and-real-world-evidence-support-regulatory-decision-making-drug.
64. US Food and Drug Administration. Data Standards for Drug and Biological Product Submissions Containing Real-World Data. US FDA Draft Guidance for Industry. October 2021. Accessed January 2023. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/data-standards-drug-and-biological-product-submissions-containing-real-world-data
65. Rosas SR, Kane M. Quality and rigor of the concept mapping methodology: a pooled study analysis. Eval Program Plann. 2012; 35(2): 236–245. DOI: http://doi.org/10.1016/j.evalprogplan.2011.10.003
66. Block LJ, Wong ST, Handfield S, Hart R, Currie LM. Comparison of terminology mapping methods for nursing wound care knowledge representation. Int J Med Inform. 2021; 153: 104539. DOI: http://doi.org/10.1016/j.ijmedinf.2021.104539
67. Saitwal H, Qing D, Jones S, Bernstam EV, Chute CG, Johnson TR. Cross-terminology mapping challenges: a demonstration using medication terminological systems [published correction appears in J Biomed Inform. 2012 Dec; 45(6): 1217]. J Biomed Inform. 2012; 45(4): 613–625. DOI: http://doi.org/10.1016/j.jbi.2012.06.005