Introduction
Clinical Data Interchange Standards Consortium (CDISC) standards and associated metadata are required in clinical trial submission deliverables.1,2 Many clinical trial data management tools, vendors, and sponsors have gone to great lengths to verify and ensure that submission documents adhere to those standards.3 Two key submission deliverables are the study data tabulation model (SDTM) annotated case report form (CRF) and the blank CRF. In this article, the author presents a simple example of a one-page CRF containing a single question collecting Fitzpatrick Skin Classification.4 The principles described are not limited to this simple example but apply to any CRF page represented in an operational data model extensible markup language (ODM-XML) file. In a submission following the CDISC SDTM standard, and associated US Food and Drug Administration (FDA) and Japan Pharmaceuticals and Medical Devices Agency (PMDA) nomenclature, the SDTM annotated CRF has the fixed name ‘acrf.pdf’ and the blank CRF has the fixed name ‘bcrf.pdf’, and they are often created manually, almost as an afterthought, despite being essential in ensuring traceability in a submission package.
ODM-XML is the primary CDISC specification document for the CRF and is a pure XML metadata specification documenting the collection of clinical data.5 The ODM-XML can contain clinical data as well; however, representation of clinical data in the ODM standard is not described in this paper. The ODM-XML standard contains all the information necessary to create both the required submission documents regarding blank and annotated CRF pages and can be used as a specification for a subcontractor (also known as a contract research organization [CRO]) engaged in the creation of CRFs in any electronic data capture (EDC) system. The ODM-XML standard is system and vendor agnostic, and easily expandable for specific system needs.6
The author proposes that by using metadata as specifications rather than documentation, and by creating submission deliverables from the metadata through validated computer code, alignment of deliverables and specifications can be assured, and most control and verification activities can be reduced. Data and documents generated under guidance of metadata can be generated much faster and much more accurately, allowing the metadata to play a double role as both specification and documentation without change. This dual role of the metadata positively influences the timelines and cost of study setup and the creation of submission deliverables. The original motivation for the author to create a style sheet was to avoid manually moving boxes within a portable document format (PDF) document. This task is best left to a computer and reduces the need to find and update page references for the define-xml.
Current State
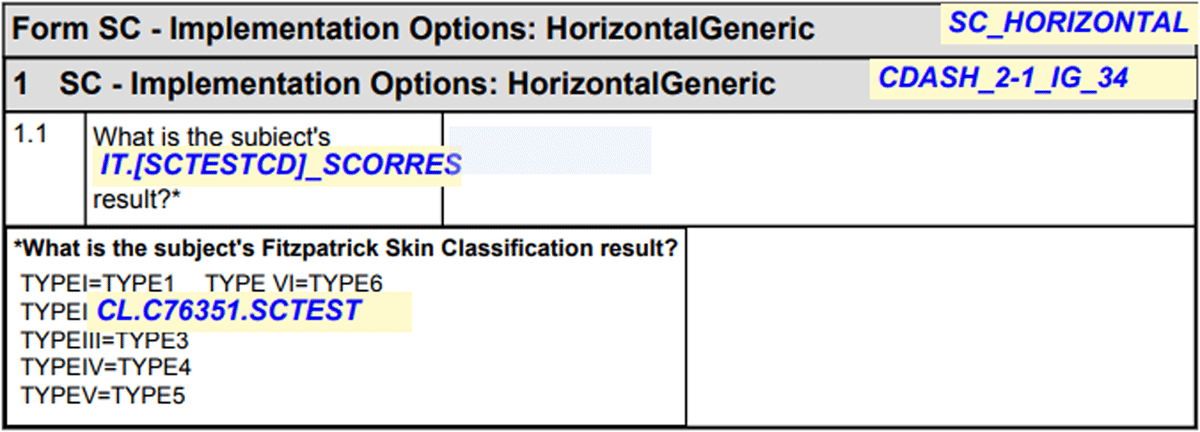
Currently, documents describing the CRF are expected in PDF. The general assumption is that they must be created using PDF creation software. This assumption is further strengthened by the common habit of creating the documents on an ad hoc basis for each clinical study and by the common, yet false, notion that CRF annotations must be displayed as text boxes in a separate layer on top of the CRF visual representation. An example of this approach can be seen when downloading sample annotated CRFs from the CDISC eCRF portal.4 These CRF pages are annotated using clinical data acquisition standards harmonization (CDASH) standards, as shown in Figure 1. SDTM annotations are normally created in the same style.
The example in Figure 1 was generated using Formedix-On, a common software package used in the creation of clinical trial documentation and is a typical example demonstrating issues originating from the habit of creating CRF pages manually using PDF creation software.
This layout has several problems:
The page displays a CRF page section (1 SC – Implementation Options: HorizontalGeneric) even when the section does not add any real value to the page. This section can hold information about display options (i.e., horizontal versus vertical layout) or annotation datasets. The orientation can be displayed in many ways, graphically, as design notes or implementation notes. The datasets impose a much more significant problem, as it imposes an unnecessary binding between the CRF pages structural layout and the annotations. This issue will be discussed in further depth in a subsequent section of this paper.
The annotations partly overlay the text of the various parts of the CRF page, obscuring features of the CRF. The traditional remedy for this problem is to have users manually move boxes around the PDF, which often changes the layout of very text dense CRF designs.
Color coding, fonts, boxes, and other formatting characteristics are necessary to distinguish annotations from the CRF content. This may support readability but may also lead to ambiguities, when annotations overlap or compete for space on very dense CRF pages.
In the example, annotations are in various places relative to the object being annotated. In cases where the annotation text is bigger than the CRF contents, the annotation spills over into other segments of the annotation, which may lead to ambiguities.
The relationship between the question and the controlled terminology may be easily detectable in a small example, but for pages having many questions referring to different, or even variations of the same, controlled terminology, it quickly becomes difficult to keep track of which controlled terminology refers to which question or requires the page layout be dictated by the annotations.
Proposed Future State
The aforementioned challenges can be solved using available software that renders the CRF and annotations based on form and data definition metadata. The rendered blank and annotated CRFs show exactly the same information as manually annotated PDFs and leave plenty of room for detailed annotations without the need for manually manipulating the CRF in PDF creation and formatting software.
The author proposes using ODM-XML to create the CRF-related submission deliverables using validated computer code, thus eliminating the bulk of controls and verification for a study. Vendors such as Formedix, Nurocor, Sycamore, and Entimo are examples of metadata repositories containing the necessary business logic to create ODM-XML files defining CRF pages.
By applying a style sheet (i.e., a special translating style sheet available on GitHub; https://github.com/jmangori/CDISC-ODM-XML-CRF-SDTM-Annotations)7 to the ODM-XML, it can be displayed as either an SDTM annotated CRF, a blank CRF, or even a specification document in any modern browser (e.g., Google Chrome, Microsoft Edge, Mozilla Firefox). PDF documents can be created via a simple print to PDF, a standard functionality supported by all browsers. The same style sheet can be applied several times varying only in a specified parameter, guaranteeing the synchronicity of the documents as they originate from the same source.
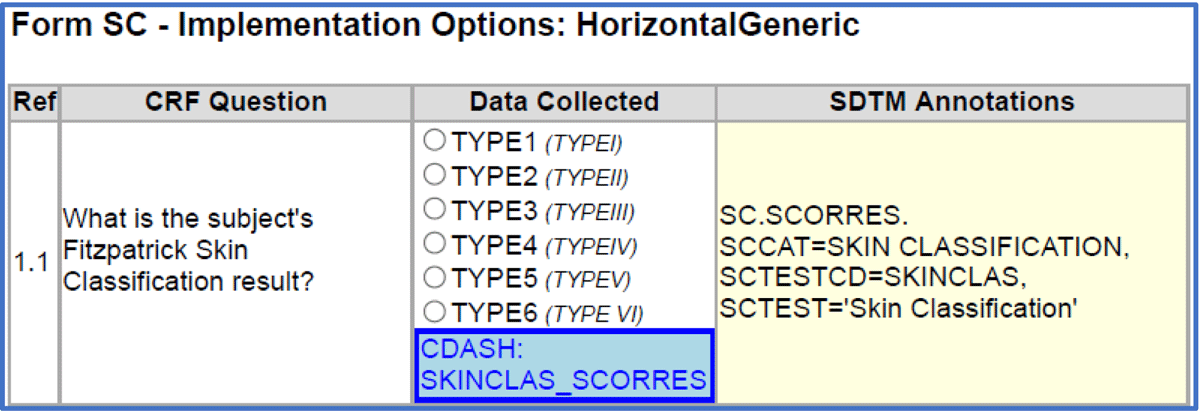
The style sheet solves the issues mentioned above while providing a method of creating the visualization of the CRF automatically without the need for human intervention. Figure 2 contains an example, also from the Fitzpatrick Skin Classification CRF, shown below.
The layout in Figure 2 is a visualization of the same ODM-XML origin as the example in Figure 1. The only difference being that the annotations are SDTM in addition to CDASH. This layout depicts several suggested solutions to the issues noted in the previous layout:
This rendering does not show implementation and design options, as they do not add any substantive value to understanding the CRF layout and the data capture.
Annotations never overlap any of the CRF text, as the SDTM annotations are displayed in a separate column as an integral part of the line defining the CRF question in its entirety.
Color coding is purely for enhanced readability, chosen to aid familiarity for the many professionals trained in CRF visualizations resembling Figure 1.
Annotations are always shown at fixed locations: CDASH annotations below the controlled terminology and SDTM annotations, including transformations, in a separate column. The browser will simply adjust the size of the cells in the table rows, accommodating for variable amounts of contents to fit any screen size.
The relationship between question text, data collected, controlled terminology, and location of data items in SDTM is always easily detectable regardless of the size and number of objects, as each question is displayed as one row of data. Additional guidance text, implementation notes, and so forth can be added in any cell without compromising traceability. Furthermore, these and other supporting texts can be added below the CRF page using the reference number as exactly that, a reference.
The immediate logical argument against this arrangement of CRF text and annotations in CDASH, SDTM, or other commonly used formats is that it takes space from the CRF layout itself by claiming an extra column compared to Figure 1. This argument overlooks that the SDTM annotated CRF is not a definition of the CRF layout. The role of visualizing layout belongs to the blank CRF. The blank CRF visualizing the FitzPatrick Skin Classification is provided in Figure 3. The Figure 3 visualization was produced by the same ODM-XML file and the same translating style sheet, differing only in a simple parameter supplied from outside.
This separation of the blank CRF and the SDTM annotated CRF enhances the transparency by making the SDTM annotated CRF the primary transparency tool and the blank CRF a reference document in the rare cases where the SDTM annotated CRF is not enough by itself. Use cases can include long lists of controlled terminology and lengthy supporting texts.
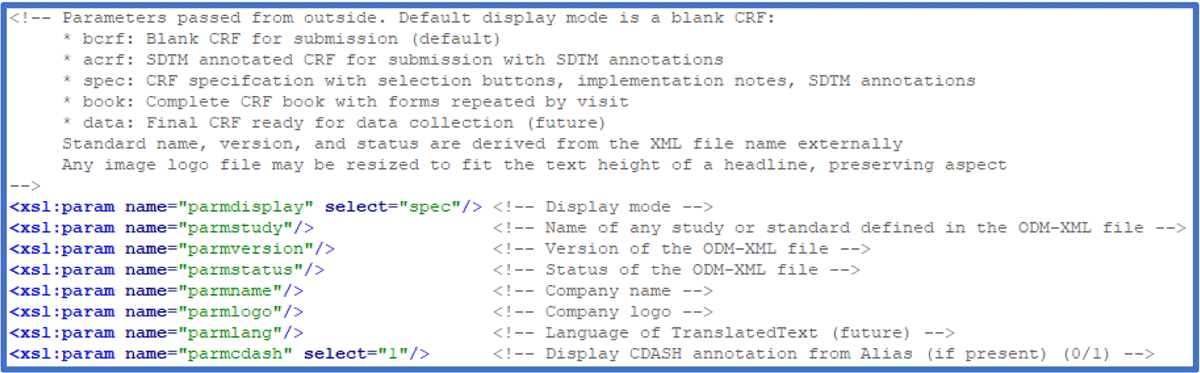
This article describes the overall principles and some of the design choices made. Recent development in browser technology has focused on security issues, including risk reduction and mitigation to prevent injection of malignant code into web pages. One consequence of this is that browsers no longer permit any web document (i.e., HTML, XML) to allow code to be run that is not part of the document itself. One solution is to write HTML code linking the ODM-XML file and the style sheet requesting the transformation of the ODM-XML to HTML guided by the translating style sheet. HTML is the ideal place determining the nature of the visualization. Figure 4 shows an excerpt of the translating style sheet defining and documenting the parameters.
A translating style sheet is a template used for converting an XML document to either another XML document, an HTML document, or other web page document types. The style sheet defines the rules for all such transformations by rearranging, adding, and subtracting data and making structural choices. As the style sheet consists of a fixed set of rules, much like a program, it is both source and deliverable agnostic.
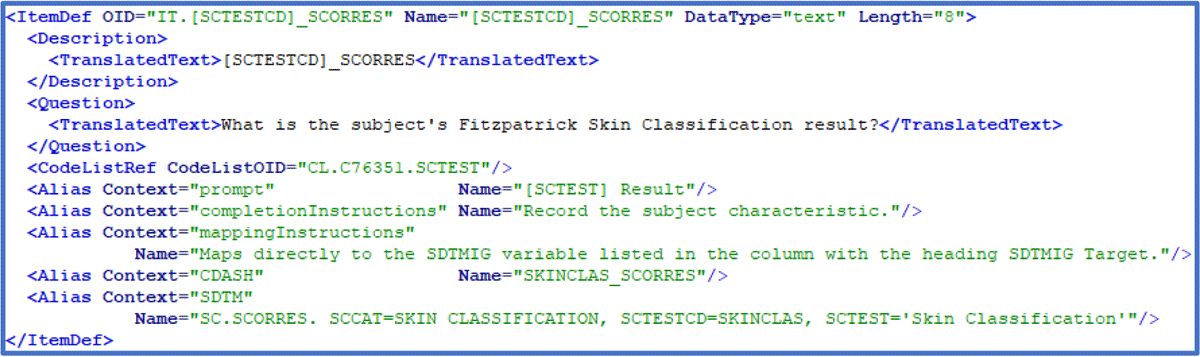
In the ODM-XML excerpt shown in Figure 5, several items are shown that are not included in either visualization of the example CRF page. The design choices within the style sheet do not display them, but different parameter settings will do so, without making any change to the ODM-XML contents. The last line of ‘Alias’ definitions in the excerpt is the SDTM annotation added by the author.
Several aspects of the metadata-driven approach bear additional mention.
The ODM-XML defines a length and a data type for the question, which may or may not impact how the answer to the question is collected as data.
The XML tag ‘ItemGroupDef’ (not shown) defines a ‘Domain’ attribute specifying which dataset the collected data should be stored in. As ‘ItemGroupDef’ in ODM-XML is the definition of sections on the CRF page, this is exactly where the unfortunate binding of the datasets and the CRF page sections is located. By ignoring the ‘Domain’ attribute and making the dataset name a prefix to the variable name in the ‘Alias’ SDTM annotation (SC.SCORRES), the dataset name is clearly and unambiguously specified exactly where it is relevant, independent of the way the CRF page is structured into sections. As all ‘ItemDef’ questions belong to one or more ‘ItemGroupDef’ sections via a reference, the ‘Domain’ attribute imposes the restriction that all questions in a section must be annotated to the same dataset. Furthermore, as the vast majority of SDTM variable names are unique across domains (and thus datasets), there is little point in trying to reuse questions across sections in one-to-many relationships.
The use of several lines of ‘Alias’ to the same question demonstrates the principle of support for multiple purposes. Support for different vendor electronic data capture (EDC) systems can be added in this way.
The syntax of the text within the ‘Alias’ having the attribute ‘Context=“SDTM”’ is an example of structured text resembling computer code, and more formal than pseudo code. The strict use of periods and commas allows the text to be parsed for extraction of definitions and may also serve as directives for formatting the text. Figure 2 shows an example in the SDTM annotations of the character sequence ‘period blank’ immediately following SC.SCORRES being visualized as a line break.
The rich ‘Alias’ SDTM annotation does not overlay the contents of the ‘Alias’ having ‘Context=“mappingInstructions”’, as the former relates to SDTM annotations and the latter to CDASH annotations.
Interlinking Documents
Define-xml8 is a well-established standard for documenting submission data. The standard way of displaying define-xml is through a similar translating style sheet provided by CDISC, containing links to targets in the SDTM annotated CRF (acrf.pdf) document. The translating style sheet used to visualize the SDTM annotated CRF from the ODM-XML creates targets for all forementioned links. When placing the define-xml document in the same folder as the acrf.pdf document, a click on a link specifying a CRF origin for any data item in define-xml opens the acrf.pdf document at the precise location of the same data item where it is collected on the CRF page. The only prerequisite for enabling this feature is to define the targets for CRF origins as named destinations in define-xml, eliminating the entire process of identifying page numbers on the CRF and creating the targets manually. Define-xml has included this feature since 2014,9 with later versions of the CDISC-supplied style sheet enabling links to named destinations.
Conclusion
Creating and validating style sheets for ODM-XML as outlined in this paper and updated as new versions of ODM-XML become available will eliminate labor intense, manual tasks during the setup of a study and the creation of a clinical trial submission package. These activities, particularly creation of the submission package, directly affect the time to market for the product being submitted.
As the style sheet accepts several external parameters, the content can be controlled when producing documents with (aCRF, CDASH, SDTM or other) or without annotations (bCRF) without changing the code of either the ODM-XML file or the style sheet.
The introduction of a style sheet for visualization of the variations of the CRF pages from ODM-XML saves time and reduces tedious and error-prone activities for users while greatly reducing cross reference checking between metadata and the generated documents, and increases transparency of submission packages supporting regulatory review. This approach (use of ODM-XML via the translating style sheet to render blank and annotated CRFs) is offered as a more efficient way to ensure transparency and to provide machine and human readable documentation to the benefit of clinical study stakeholders.
Competing Interests
The author has no competing interests to declare.
References
1. United States Food and Drug Administration (FDA). eCTD Technical Conformance Guide. Updated February 25, 2022. Accessed May 6, 2022. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/ectd-technical-conformance-guide.
2. Ando Y. Pharmaceuticals and Medical Devices Agency (PMDA). Technical Guide for Electronic Data Submission in Japan. Updated January 24, 2019. Accessed May 6, 2022. https://www.pmda.go.jp/english/review-services/reviews/0002.html.
3. Pinnacle 21 by Certara. Solutions. Accessed May 6, 2022. https://www.pinnacle21.com/products/validation.
4. Clinical Data Interchange Standards Consortium (CDISC). eCRF Portal, Subject Characteristics CDASHIG v2.1. Accessed May 6, 2022. https://www.cdisc.org/sites/default/files/kb/ecrf/SC_eCRF_Package.zip.
5. Clinical Data Interchange Standards Consortium (CDISC). Specification for the Operational Data Model (ODM), Version 1.3.2 Production. Accessed May 6, 2022. https://www.cdisc.org/system/files/members/standard/foundational/odm/odm1_3_2.zip.
6. Clinical Data Interchange Standards Consortium (CDISC). Specification for the Operational Data Model (ODM), Version 1.3.2 Production, Sections: 1 Introduction (non-normative), 2.3 System Conformity, 2.4 Vendor Extensions. Accessed May 6, 2022. https://www.cdisc.org/system/files/members/standard/foundational/odm/odm1_3_2.zip.
7. Iversen J. jmangori/CDISC-ODM-XML-CRF-SDTM-Annotations. Accessed October 3, 2022. https://github.com/jmangori/CDISC-ODM-XML-CRF-SDTM-Annotations
8. Clinical Data Interchange Standards Consortium (CDISC). Define-xml version 2.0 definition. Accessed May 6, 2022. https://www.cdisc.org/standards/data-exchange/define-xml.
9. Clinical Data Interchange Standards Consortium (CDISC). Define-xml version 2.0 definition. Accessed May 6, 2022. https://www.cdisc.org/standards/data-exchange/define-xml#standard__versions.